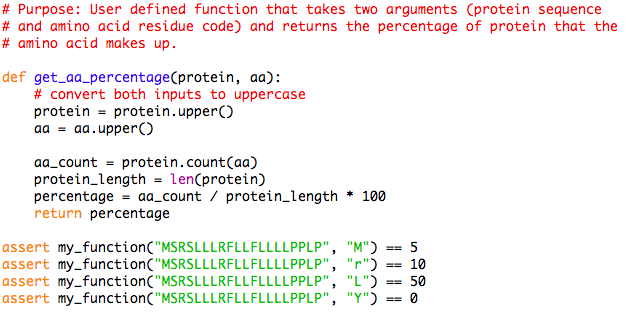
Python script to recode an amino acid sequence into generic format for... | Download Scientific Diagram

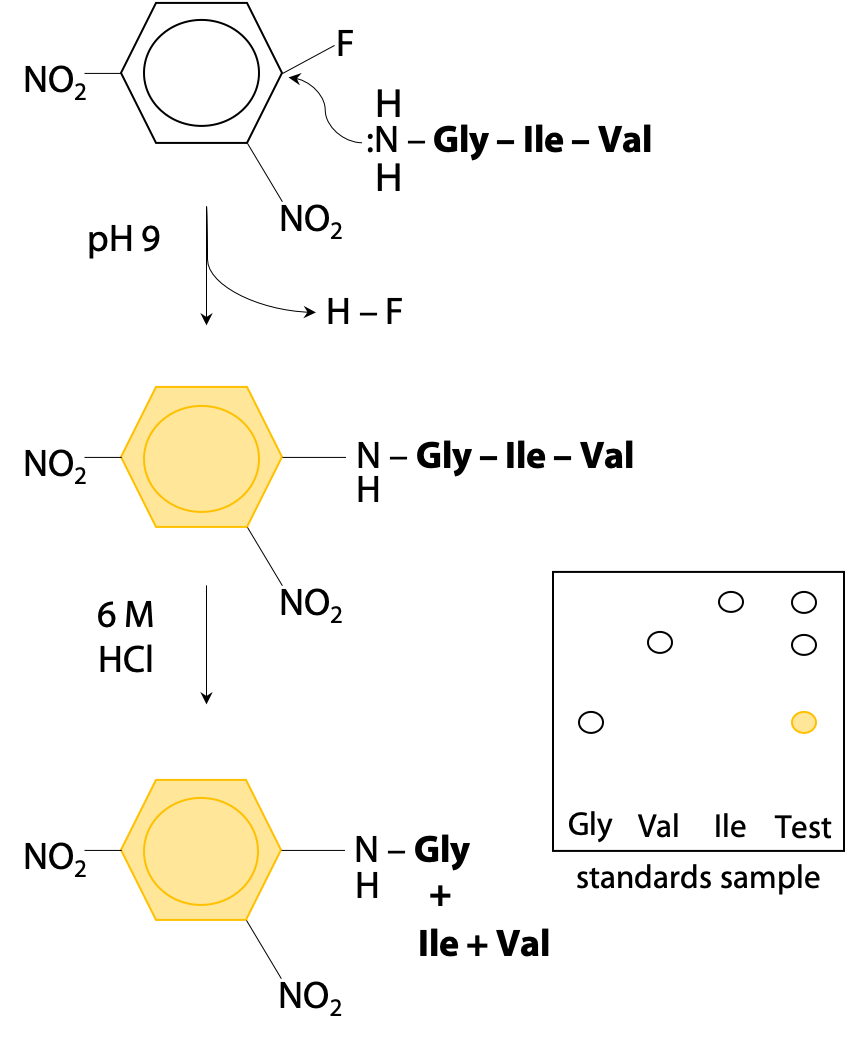
Data acquisition and database search. For a given experimental MS/MS... | Download Scientific Diagram
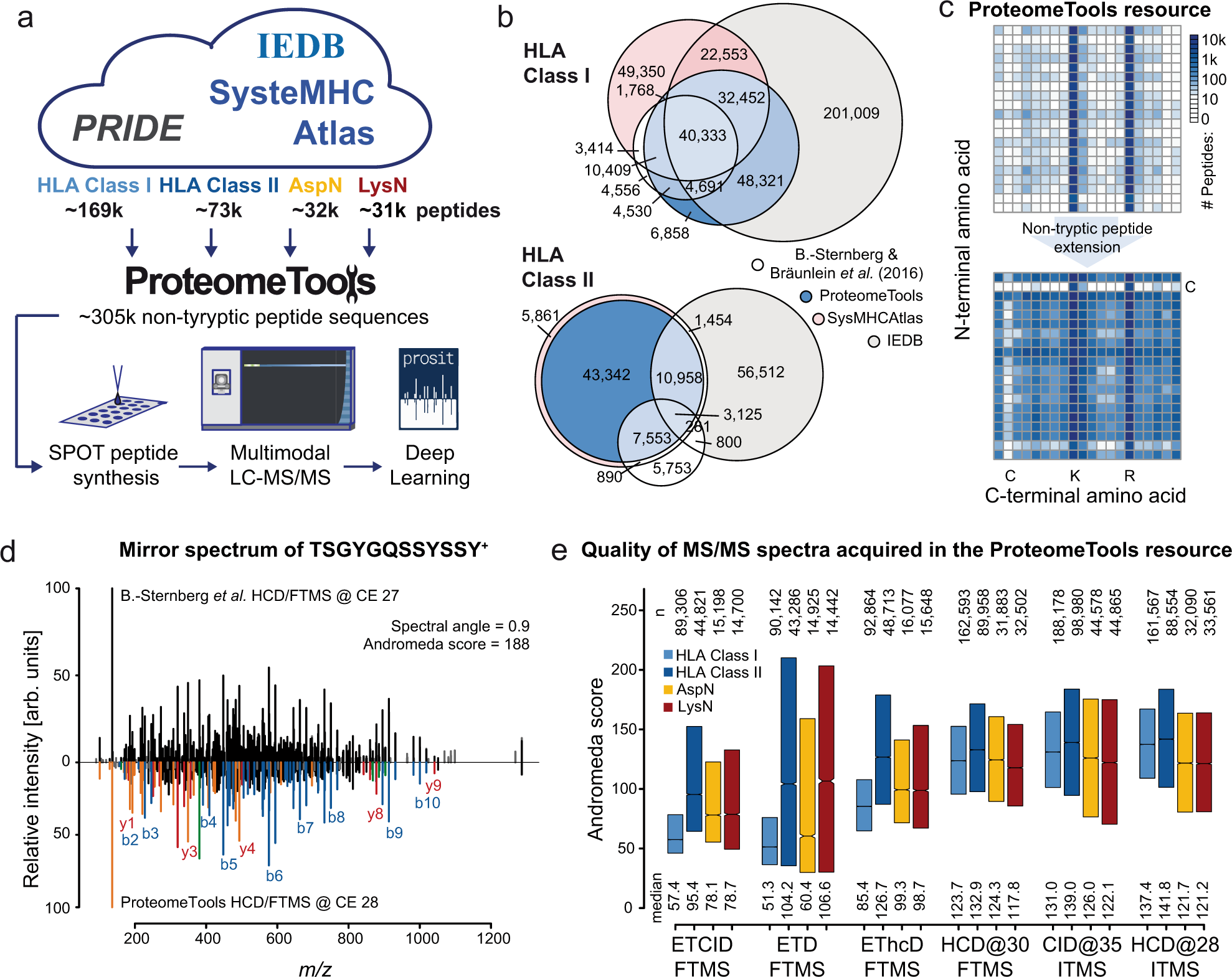
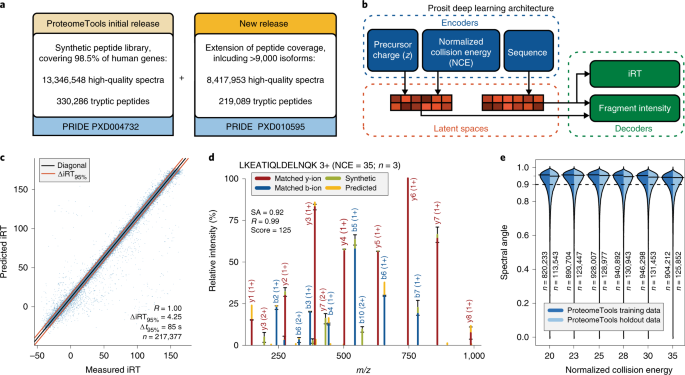
Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications
ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis | PLOS Computational Biology
Functional expression of diverse post-translational peptide-modifying enzymes in Escherichia coli under uniform expression and purification conditions | PLOS ONE

Protein structure, amino acid composition and sequence determine proteome vulnerability to oxidation‐induced damage | The EMBO Journal
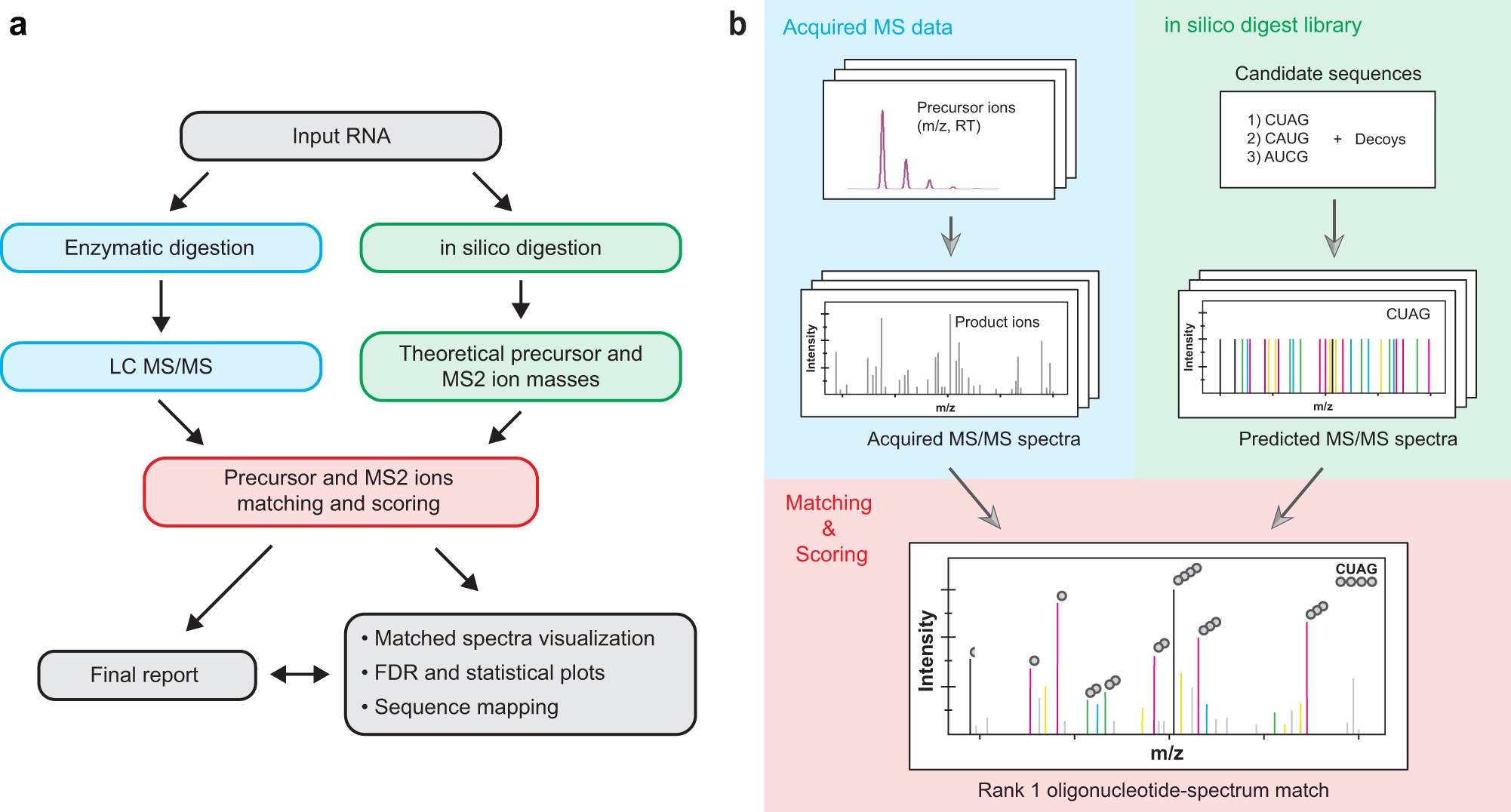
Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry | Nature Communications
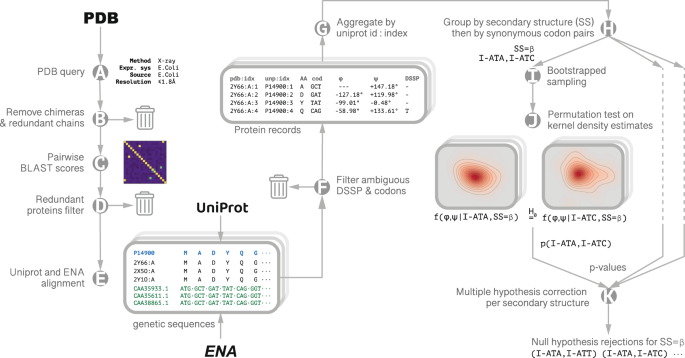
Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon | Nature Communications
GitHub - efr-essakhan/pepfeature: A Python package that has functions for featue calculation of protein sequences. Features that are calculated by this package are niched for Epitope prediction.